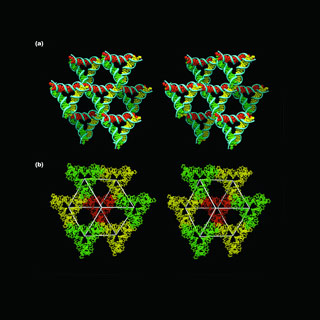
Scientists, counting those who were occupied in this research, have apparently earlier designed and built crystal structures. But these compositions were supposedly two-dimensional which may mean that their axes may be on a single plane and may not be the most absolute depiction of crystals.
To tackle this drawback, the research team, sought to design and build three-dimensional DNA crystals, a procedure that may need important spatial control of the 3D structure of matter. Led by NYU Chemistry Professor Nadrian Seeman researchers from Purdue University’s Department of Chemistry and the Argonne National Laboratory in Illinois were also included in the research.
To do this, the researchers apparently formed DNA crystals by making synthetic sequences of DNA that may have the capability to self-assemble into a sequence of 3D triangle-like motifs. The formation of the crystals was supposedly reliant on putting ‘sticky ends’ i.e. minute unified sequences on every end of the motif that apparently fastens to other molecules and position them in a set order and orientation. The composition of these sticky ends may let the motifs join to each other in a planned manner.
Seeman and his colleagues claimed to have formerly produced crystals via this route. Nevertheless, as these crystals self-assembled on the same plane, they appeared to be two-dimensional in composition. The researchers prolonged on the previous efforts by taking benefit of DNA’s double-helix structure to supposedly form 3D crystals. The 2D crystals may be extremely tiny, an approximate 1/1000th of a millimeter, but the 3D crystals are supposedly between 1/4 and 1 millimeter, which may be seen by the naked eye.
DNA’s double helices may take shape when single strands of DNA, apparently each comprising of about four molecular components known as bases. These may be connected to backbone, self-assembled by corresponding up their bases. The researchers added sticky ends to these double helices, which may outline single-stranded overhangs to every double helix. These overhanging sticky ends attach together to tie two double helices. This is supposedly a general method used by genetic engineers, who apparently use it on a much superior level. By connecting together multiple helices through single-stranded sticky ends, the researchers claimed to form a lattice-like structure that broadens in six diverse directions, thus apparently yielding a 3D crystal.
Seeman commented, “With this technique we can organize more matter and work with it in many more ways than we could with 2D crystals.â€
A potential avenue for the use of this approach may be in nanoelectronics, via components which may not be bigger than single molecules. Presently, such products could be constructed with 2D components. Given the improved flexibility that 3D components may yield, manufacturers could assemble parts that may be smaller and closer jointly as well as supposedly more classy in design.
The researchers also anticipated that they may be able to organize biological macromolecules by joining them to these crystals. This may assist in the development of drugs as macromolecules arranged in crystals may be pictured by a procedure called as X-ray crystallography. By adding drugs to these crystals, their communication with these biological components may be pictured.
X-ray diffraction data could have been collected from DNA crystals and their iodinated derivatives on beamlines X6A and X25 at the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory in Upton, New York and on beamline ID19 at the Structural Biology Center at Advanced Photon Source in Argonne, Illinois.
Bob Sweet, a biophysicist and the leader of the group who run NSLS beamline X25 observed that it is one of the neatest structures, he has seen in years. As per him, it really connects biotechnology to nanotechnology.
This research was published in the latest issue of the journal Nature
