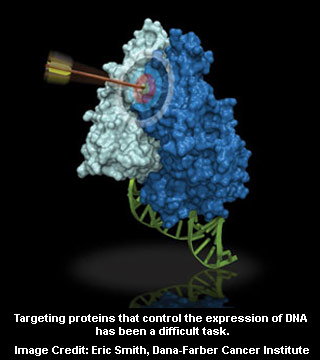
The uncontrollable cell growth also known as transcription factors appear to sit on DNA and turn genes on and off. This transcription factor apparently defines cancer results from runaway growth genes activated by regulator proteins. Moreover, they seem to be frequently mutated in cancer.
Supposedly, the authors developed this by combining advances in chemistry, chemical biology and genomics. Bradner was believed to have examined patients with advanced blood cancers like T-cell acute lymphoblastic leukemia (T-ALL).
“Unfortunately, by the time I see these patients, they’ve already failed to respond to conventional therapy. It’s in these patients where new therapies are most urgently needed,†says lead researcher, Jay Bradner, also a chemical biologist and oncologist at the Dana-Farber Cancer Institute and an associate member of the Broad Institute of MIT and Harvard.
Jon Aster, an associate professor in the Department of Pathology at Harvard Medical School and co-researcher was noted to have discovered in 2004 that approximately 50% of patients with T-ALL have mutations in a gene called NOTCH.
It was originally named NOTCH because for its effect on the notched wings of fruit flies when altered. NOTCH may be able to assist cells throughout the body in order to make decisions about what to become, e.g. to become T-cell or B-cell. If NOTCH is mutated and becomes too active, it could drive cancer. Since the NOTCH protein appears to be a transcription factor, there were no clear ways to target it molecularly.
“Proteins that regulate gene activity usually come together like two noodles, with expansive interfaces, so getting a molecule to bind just right and block the interface is a real challenge,†elucidates Bradner.
Targeting proteins with drugs or molecules may usually require some kind of groove or pocket in the target. A small molecule could perhaps nestle into this groove and block the protein from doing its job. Moreover, transcription factors seem to have such large, smooth interfaces that they are often termed ‘undruggable,’ at least via conventional techniques of screening for drugs claim experts.
In 2005, Aster, a co-author of the new research was believed to have uncovered the crystal structure that NOTCH forms when it partners with DNA and two other proteins. For the foremost time, researchers and physicians seem to be able to view the shape of this gene regulatory team and visualize how the molecules worked in concert to flip a genetic switch and drive cancer.
Interestingly, it seems to have revealed a long, spring-like section of one protein, known as an alpha-helix that sits in a groove in the NOTCH protein. Bradner speculated that if something could be designed to fit in the groove and prevent this protein from teaming up with NOTCH thereby essentially blocking its function.
The alpha-helix is known to be a common shape formed by proteins and chemists can build it in the lab. However, a short section of alpha-helix tends to unravel thereby reducing its ability to bind a target. Apparently, Bradner required a way to hold back the alpha-helix into a spring-like shape.
Not only the stapling method seems to be tailored to alpha-helices, but it accomplishes feats impossible with other molecules. Just adding up this very simple chemical modification may cause a bunch of things to happen all immediately.
Furthermore, the method doesn’t simply impose the helical shape between ends of the ‘staple,’ it apparently holds the entire peptide tightly in shape. This, in turn, appears to prevent cellular enzymes from breaking down the proteins, a major way of peptide inactivation.
Also, stapled proteins stay active longer in the body thereby providing them a greater chance of performing their task. Most considerably, stapling is known to allow the peptides to enter cells. This appears to be magical, because it solves the biggest problem the researchers face with unmodified peptides.
Ray Moellering, a graduate student in Harvard University’s Department of Chemistry and Chemical Biology created six overlapping stapled alpha-helices that seem to have covered the whole length of one of NOTCH’s partner proteins. Besides, one peptide, which they called SAHM1, was particularly promising.
They were believed to have produced a fluorescent version of SAHM1 and proved that it did in fact penetrate cells which were often a challenge for peptides. Allegedly, the peptide killed human cancer cells driven by NOTCH, and left alone those not driven by NOTCH. This seems to be a promising result.
By means of gene expression microarrays and the Broad’s Gene Set Expression Analysis (GSEA) software, the researchers found that the peptide entirely seems to shut down the NOTCH target genes, the very genes that cause cancerous cell growth.
Recognizing that SAHM1 could achieve such a feat i.e. inhibition of a transcription factor in cells, the researchers wondered if the peptide they had planned to reveal where to put a drug may be a drug itself. For the purpose of testing this theory, they collaborated with other researchers to develop a model of T-ALL in mice.
Through luminescent firefly genes and bone marrow transplantation, they appear to have increased a fleet of mice with ‘glowing’ tumors driven by the NOTCH mutation. The researchers found that in all the mice treated with SAHM1, NOTCH seem to have inhibited and tumors reduced in size.
A directed inhibitor of NOTCH that works in humans could be helpful for treating not only T-ALL, but other cancers like ovarian, breast, and lung cancer, as well as other medical conditions such as atherosclerosis and immune diseases.
They are currently building stapled peptides that inhibit the other three versions of NOTCH. They are also working to target other transcription factors, many thought to be undruggable, with stapled peptides. Though the research is not ready for use in patients, it may have implications beyond developing a new research tool and prototype drug for targeting NOTCH.
Moreover, it seems to provide a generic method for targeting other transcription factors, a previously unachievable goal. Researchers anticipate that scientists will make use of this template strategy to create powerful tools for studying gene regulation throughout cell biology.
The findings of the research have been published in the journal, Nature.
