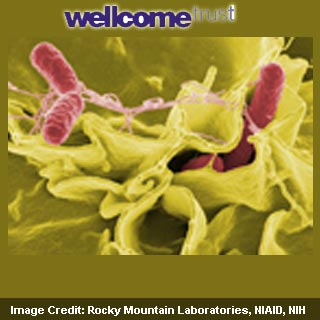
The researchers describe that genetic changes appear to have transformed an earlier mild salmonella infection into a new antibiotic-resistant form of the disease. Known to be ST313, this new form of the strain seems to be better at adapting to affect humans as compared to the more common Salmonella Typhimurium form. It appears to be resistant to several commonly used antibiotics and could spread from person-to-person. In many parts of sub-Saharan Africa, it supposedly infects vulnerable children and adults, consequently leading to death in nearly one of four cases.
Diarrhoea could be caused by the normal ST313 form of S. Typhimurium and is apparently fatal in rare cases. Originally scientists were of the opinion that the strain was passive in action and could take advantage of the weakened immune system. Thus it was particularly thought to infect people suffering from HIV, malaria, malnutrition or anemia. However, the new analysis reveals that it could be more invasive and seems to have evolved into a very dangerous and drug-resistant form.
“This is a new form of pathogen,” mentioned Dr Robert Kingsley from the Sanger Institute. “It seems to have evolved to acquire a unique armoury that allows it to infect vulnerable children and adults efficiently in some African regions.”
Starting from 2002 in an 18-month period, multidrug-resistant ST313 appeared to represent 95 per cent of S. Typhimurium isolates recognized in Africa. As part of the research, the experts examined about 50 samples of ST313 DNA that were extracted from blood samples of African patients. These individuals were known to have severe symptoms of infection and also suffered from HIV, malaria, malnutrition or anemia. Their combined genetic sequence from these samples was then pitted against those of strains that could cause milder disease symptoms.
“We saw genetic signatures that suggested ST313 may be adapting to humans,” remarked Dr Kingsley. “For example, this deadly strain has lost around one in 50 of the genes found in the ‘typical’ S. Typhimurium – a classic sign that it may be becoming more closely adapted to one host, in this case, humans.”
ST313 may be spreading by a new method, not seen before in S. Typhimurium proposes the research. Normally circulating among animals, the pathogen may be introduced to humans via food poisoning. It apparently has the ability to adapt quickly to its human hosts mainly because it could be passing predominantly from person to person.
“If we can understand what’s special about the types of Typhimurium that emerge and susceptible populations, we might be able to predict in future where new pathogens will emerge,” said Professor Gordon Dougan from the Sanger Institute. “We may also be able to design vaccines against those pathogens.”
Researchers are hopeful of coming up with efficient vaccines by better understanding this new deadly strain.
